Introduction
This blog post is a summary of a dataset with information that may be useful in current projects. And the paper is:
CaDIS: Cataract Dataset for Image Segmentation
This blog contains the following contents:
Dataset
Experiments
Problems of this dataset
Note: This blog represents only the Information extraction.
Dataset
Annotation process
Some points to note in the annotation:
The number of pixels for each class is greater than 50;
Annotation errors may originate from: blurred boundaries caused by motion. In addition, specular reflections may also lead to inaccurate boundary delineation, especially for the instrument tips when they are inside the anatomy.
Dataset statistics
The dataset includes 36 classes: 29 surgical instrument classes, 4 anatomy classes and 3 miscellaneous classes, namely: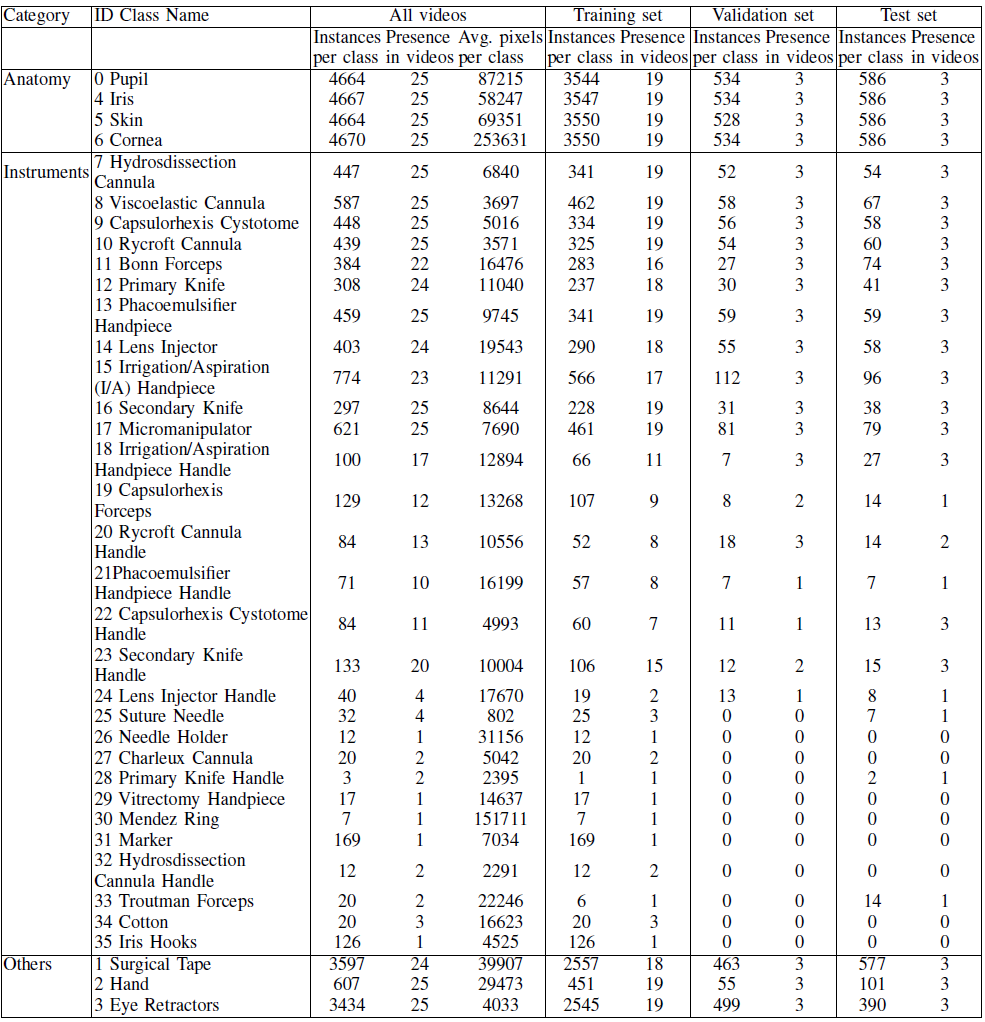
Classes are imbalanced:
The anatomy classes appear more frequently than the surgical instruments. The anatomy also covers the largest part of the scene.
The classes about “Instrument” in dataset is highly imbalanced, for example, there are 17 instrument classes appear in less than half of the videos. So, accurate instrument classification is more challenging.
Some Instruments have high similarity but completely different functions, such as “cannulas”, which is also a difficulty of this dataset.
Experiments
Three different tasks
Three different tasks are proposed in this dataset, that is, classes are merged in three different ways.
Task 1 (8 classes):
The first task is focused on differentiating between anatomy and instruments within every frame. That is, here we only have 8 classes: 4 anatomy classes, 3 miscellaneous classes and 1 classes with all Instruments:
Task 2 (17 classes):
The second task is not only focuses on anatomy, but also classifies Instruments based on appearance similarity and instrument type. That is, this task is to identify anatomical structures and also the main types of instruments that appear in the scene.
The classes are as follow:
The specific merged classes are as follows:
Cannula
- Hydrosdissection Cannula; Viscoelastic Cannula; Rycroft Cannula; Rycroft Cannula Handle; Charleux Cannula; Hydrosdissection Cannula Handle
Cap. Cystotome
- Capsulorhexis Cystotome; Capsulorhexis Cystotome Handle
Tissue Forceps
- Bonn Forceps; Troutman Forceps
Primary Knife
- Primary Knife; Primary Knife Handle
Ph. Handpiece
- Phacoemulsifier Handpiece; Phacoemulsifier Handpiece Handle
Lens Injector
- Lens Injector; Lens Injector Handle
I/A Handpiece
- Irrigation/Aspiration (I/A) Handpiece; Irrigation/Aspiration Handpiece Handle
Secondary Knife
- Secondary Knife; Secondary Knife Handle
Micromanipulator
- Micromanipulator
Cap. Forceps
- Capsulorhexis Forceps
Ignore
- Suture Needle; Needle Holder; Vitrectomy Handpiece; Mendez Ring; Marker; Cotton; Iris Hooks
Note: the instruments that only appear in the training set, cover relatively few pixels in the frame and cannot be merged with another instrument class were ignored during training.
Task 3 (25 classes)
This task builds on Task 2 for a finer classification. In task 2, we can find that each Instrument and its corresponding Handle are merged together, and this task is to separate them.
The classes are as follow:
- Ignore
- Suture Needle; Needle Holder; Charleux Cannula; Primary Knife Handle; Vitrectomy Handpiece; Mendez Ring; Marker; Hydrosdissection Cannula Handle; Troutman Forceps; Cotton; Iris Hooks
Note: The classes that do not appear in all splits and are present in less than 5 videos were ignored during training.
Models
Three models are used in the paper, namely, UNet, DeepLabV3+, UPerNet and HRNetV2. (all the models are open-source)
For training
Training dataset
The same data augmentation was applied for all model before training, they used: Each training image was normalized, flipped, randomly rotated and hue and saturation was also adjusted. The input images were downsized to 270 x 480.
No post-processing was performed.
Training configuration
pre-trainged weight
- the weights for
UPerNetandHRNetV2were initialized using pre-trained weights on ImageNet; - the weights for
DeepLabV3+was based on Pascal VOC;
- the weights for
epochs, GPU
- 100 epochs
- two NVIDIA GTX 1080 Ti GPUs
learning rate, optimizer
- the Cross Entropy loss function was used with learning rate equal to $10^{-4}$ using the Adam Optimizer.
- The $\beta_1$, $\beta_2$ and $\epsilon$ values for the Adam Optimizer
were set to 0.9, 0.999 and $10^{-8}$, which are proposed as good
Metrics
The paper used 3 metrics to evaluate the models, namely, PA, PAC, mIOU, but because of the imbalanced of the dataset and classes (that is, some classes only appear in some videos’ frame, and the number of pixel of some classes are larger than others). So, we mainly focus on mIOU.
Results
For task 1
By using different models, the results based on task 1 are as follow: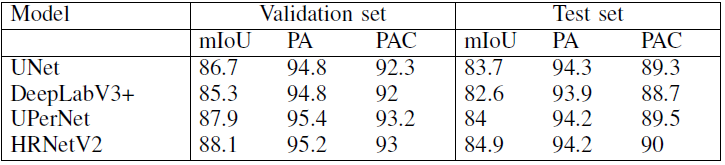
and mIOU per class is: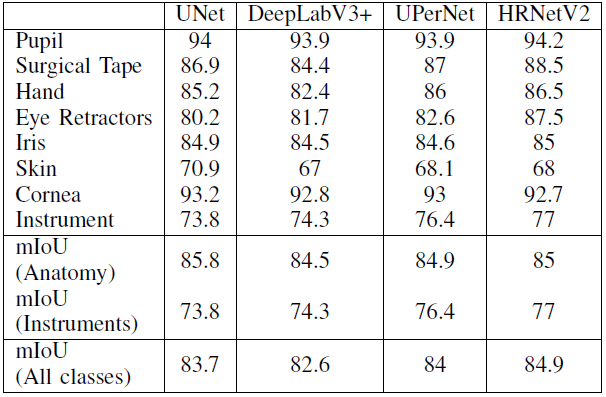
From this chart we could find that:
HRNetV2presenting the highestmIOU;DeepLabV3+presenting the lowest for both validation and test sets;- There is small differences in the mIOU between the models because the imbalance among the classes is reduced by representing all instruments with one class.
For task 2
By using different models, the results based on task 2 are as follow: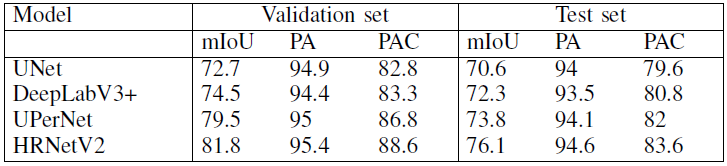
and mIOU per class is: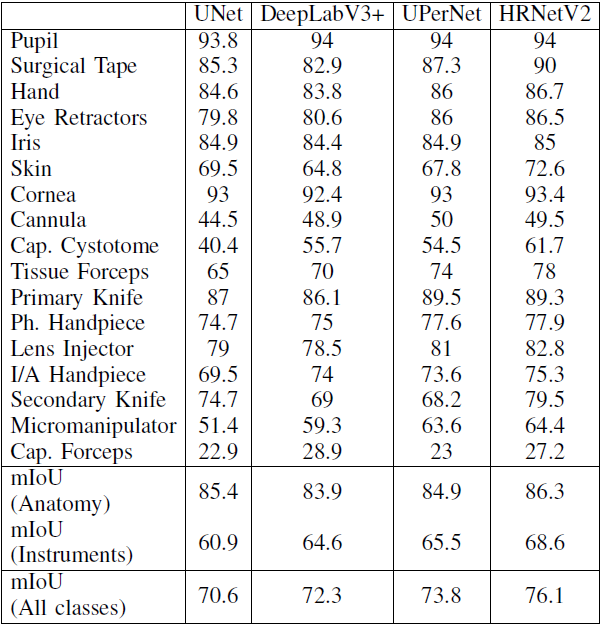
From this chart we could find that:
- The mIOU between the different models starts to differ significantly as the class Instruments starts to get imbalanced;
- For large classes, such as the anatomical classes and instrument classes that are represented by large number of pixels, have high mIOU;
- Cannula group of classes and the capsulorhexis cystotome have a relatively low mIoU, beccause these classes are represented by a small number of pixels and a large percentage of them is classified as anatomy around the boundary.
- The capsulorhexis forceps present a low mIoU for all models and this is because they are frequently misclassified as tissue forceps, These two classes could have been merged into one group, however as the capsulorhexis forceps appear in all splits with sufficient number of training images, they are represented by a separate class.
For task 3
By using different models, the results based on task 3 are as follow: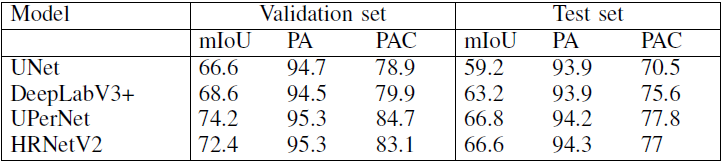
and mIOU per class is: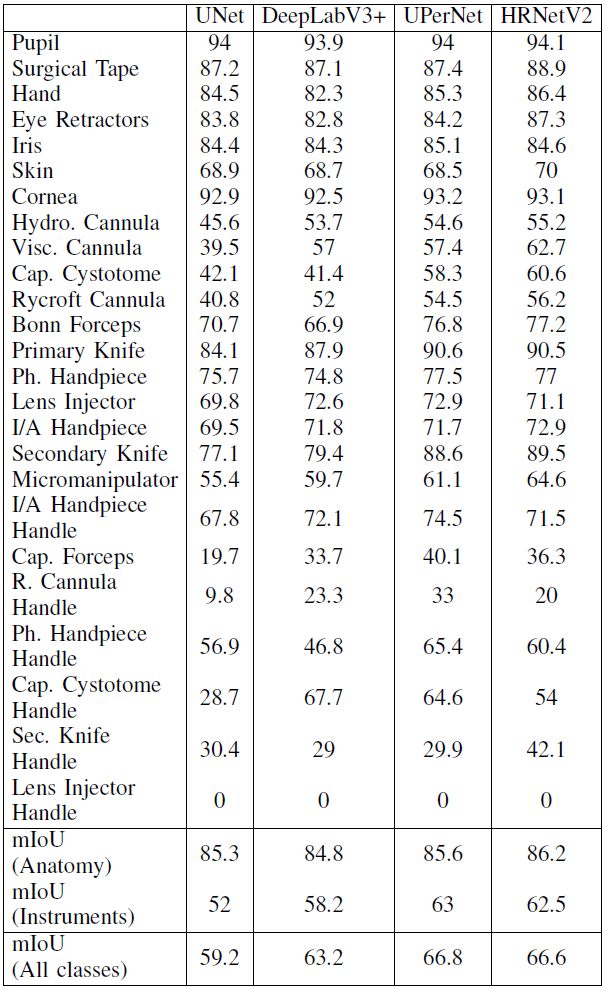
From this chart we could find that:
- As the number of classes increases, class imbalance is more evident;
Conclusion
DeepLabV3+,UPerNetandHRNetachieve higher mIOU for instrument segmentation and classification thanUNet;UNetgives a lower mIoU at instrument segmentation and classification;UPerNetandHRNethave the higher mIoU at simultaneous anatomy segmentation and instrument classification.
Additional
According to the finding of “RVIM Lab”, some annotations in the dataset are wrong, and at least 179 images were labeled wrong, and they(“RVIM Lab”) relabeled some of them. Such as the following examples: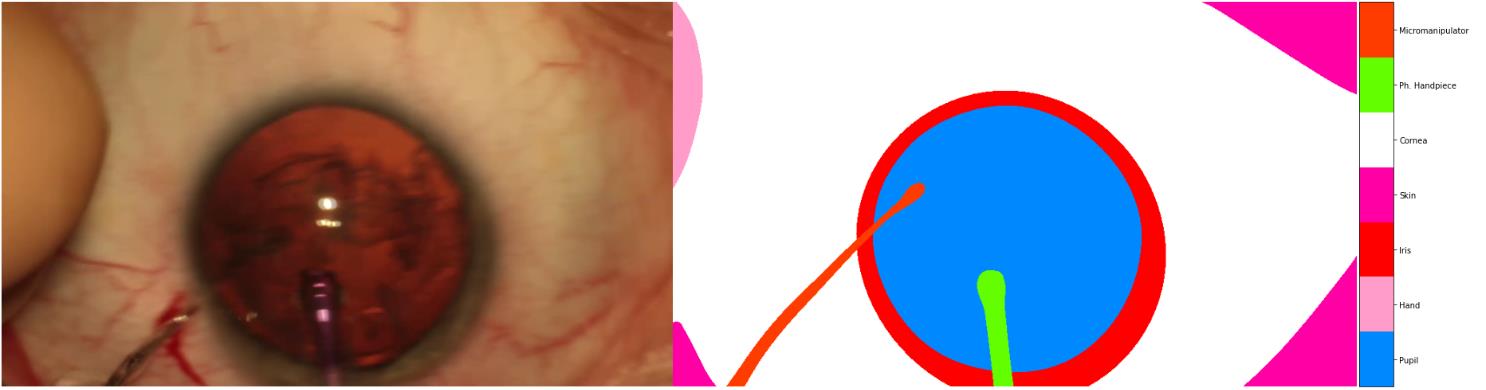
we could find that, the annotations of Hand and Micromanipulator are wrong, the correct mask should be: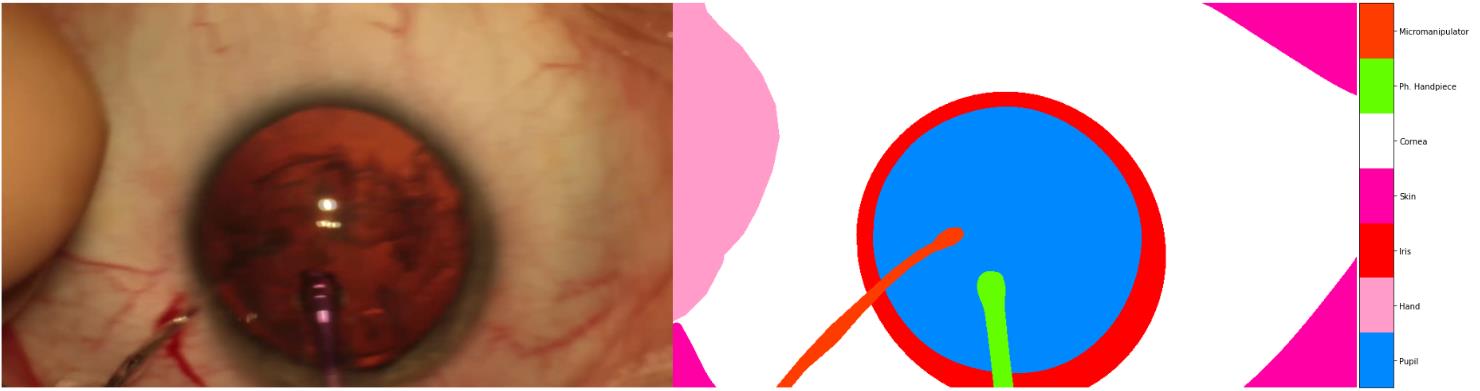
Another example: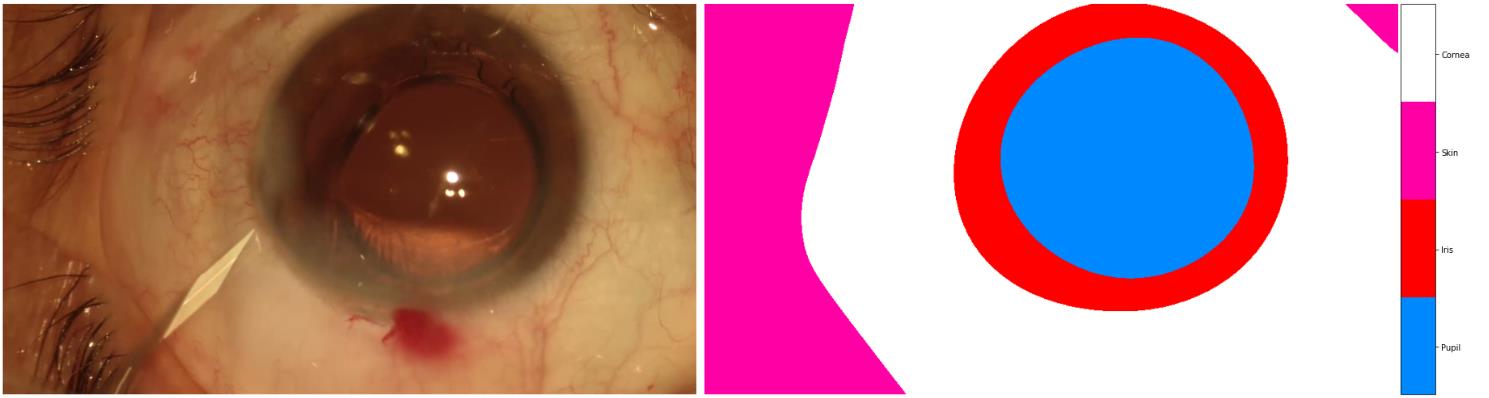
the correct one should be: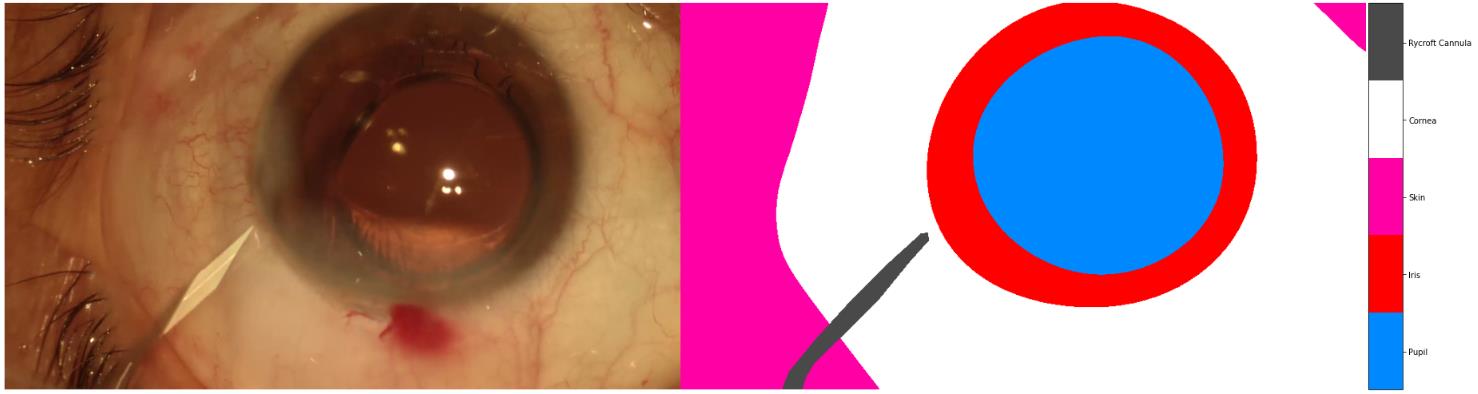
Reference:
[1] Paper: CaDIS: Cataract Dataset for Image Segmentation
[2] Paper: Effective semantic segmentation in Cataract Surgery: What matters most?